Note
Go to the end to download the full example code or to run this example in your browser via Binder
Correspondances¶
Correspondances is a mechanism to intend to continue tracking with new detection
import logging
from matplotlib import pyplot as plt
from netCDF4 import Dataset
from py_eddy_tracker import start_logger
from py_eddy_tracker.data import get_remote_demo_sample
from py_eddy_tracker.featured_tracking.area_tracker import AreaTracker
# In order to hide some warning
import py_eddy_tracker.observations.observation
from py_eddy_tracker.tracking import Correspondances
py_eddy_tracker.observations.observation._display_check_warning = False
def plot_eddy(ed):
fig = plt.figure(figsize=(10, 5))
ax = fig.add_axes([0.05, 0.03, 0.90, 0.94])
ed.plot(ax, ref=-10, marker="x")
lc = ed.display_color(ax, field=ed.time, ref=-10, intern=True)
plt.colorbar(lc).set_label("Time in Julian days (from 1950/01/01)")
ax.set_xlim(4.5, 8), ax.set_ylim(36.8, 38.3)
ax.set_aspect("equal")
ax.grid()
Get remote data, we will keep only 20 first days, get_remote_demo_sample function is only to get demo dataset, in your own case give a list of identification filename and don’t mix cyclonic and anticyclonic files.
file_objects = get_remote_demo_sample(
"eddies_med_adt_allsat_dt2018/Anticyclonic_2010_2011_2012"
)[:20]
We run a traking with a tracker which use contour overlap, on 10 first time step
c_first_run = Correspondances(
datasets=file_objects[:10], class_method=AreaTracker, virtual=4
)
start_logger().setLevel("INFO")
c_first_run.track()
start_logger().setLevel("WARNING")
with Dataset("correspondances.nc", "w") as h:
c_first_run.to_netcdf(h)
# Next step are done only to build atlas and display it
c_first_run.prepare_merging()
# We have now an eddy object
eddies_area_tracker = c_first_run.merge(raw_data=False)
eddies_area_tracker.virtual[:] = eddies_area_tracker.time == 0
eddies_area_tracker.filled_by_interpolation(eddies_area_tracker.virtual == 1)
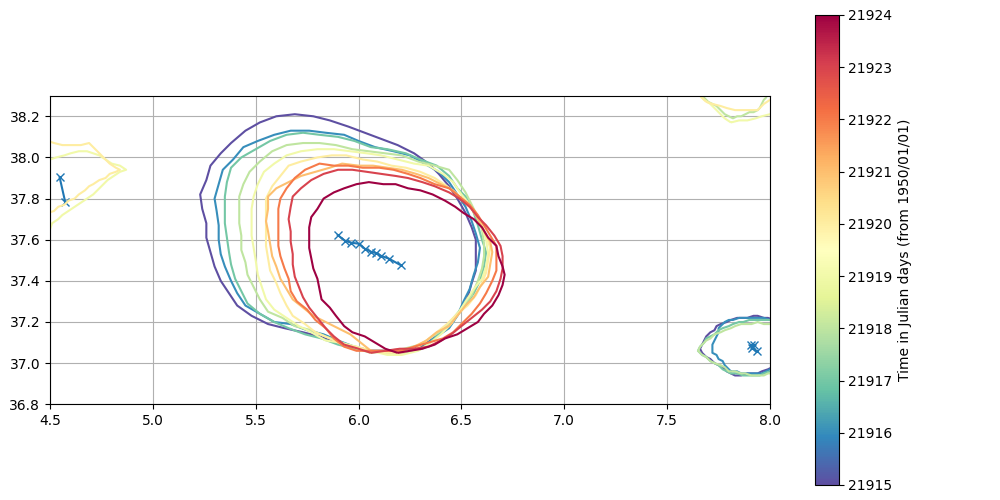
Plot from first ten days
plot_eddy(eddies_area_tracker)
Restart from previous run¶
We give all filenames, the new one and filename from previous run
c_second_run = Correspondances(
datasets=file_objects[:20],
# This parameter must be identical in each run
class_method=AreaTracker,
virtual=4,
# Previous saved correspondancs
previous_correspondance="correspondances.nc",
)
start_logger().setLevel("INFO")
c_second_run.track()
start_logger().setLevel("WARNING")
c_second_run.prepare_merging()
# We have now another eddy object
eddies_area_tracker_extend = c_second_run.merge(raw_data=False)
eddies_area_tracker_extend.virtual[:] = eddies_area_tracker_extend.time == 0
eddies_area_tracker_extend.filled_by_interpolation(
eddies_area_tracker_extend.virtual == 1
)
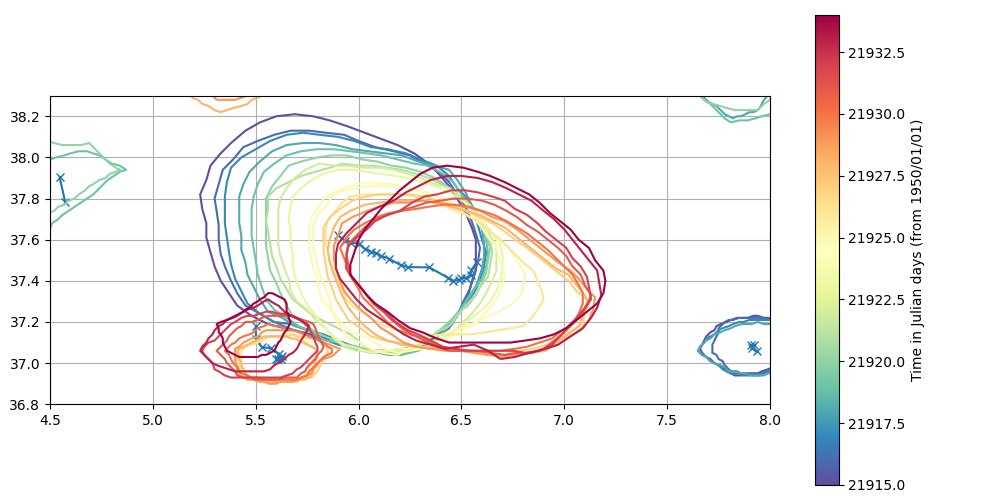
Plot with time extension
plot_eddy(eddies_area_tracker_extend)
Total running time of the script: ( 0 minutes 3.583 seconds)